Metabolomics Platform
The platform is established at the Institute for Clinical Chemistry and Laboratory Medicine of University Hospital Dresden and contributes to the mission of NCT/UCC Dresden.
Alterations in cellular metabolism are a hallmark of many cancer types. Research in this context aims to understand the role of cellular metabolism in tumorigenesis as well as developing new tumor metabolism-related therapies.
Metabolomics is the comprehensive analysis of small molecules (i.e. metabolites) in body fluids, cells, tissues and tissue biopsies. To facilitate research into cancer metabolism, the Metabolomics Platform offers metabolite analysis using complementary methods, specifically nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS).
NMR spectroscopy
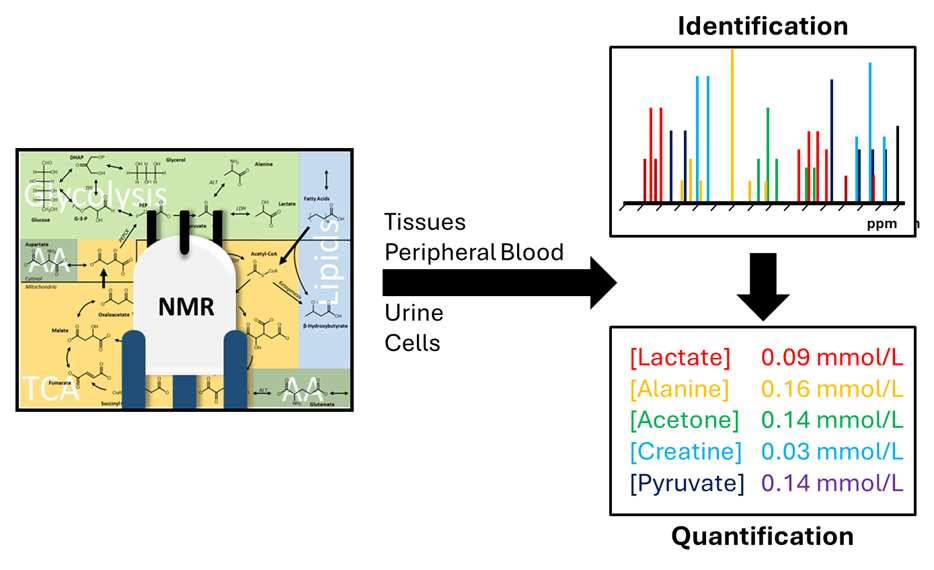
NMR spectroscopy is a robust method for quantifying metabolites. The 1H NMR-based metabolic profile allows the identification of metabolic changes in a wide range of media. NMR spectroscopy is an automated, simple (minimal sample preparation) and highly reproducible technique that can provide a quantitative fingerprint/profile with a 25-minute measurement.
Quantitative metabolomics via NMR spectroscopy (Figure 1):
- Measurement in liquid samples (quantitative analysis of up to 150 metabolites in urine; 40 metabolites in serum/plasma)
- HR-MAS (high-resolution magic angle spinning) for measurement of metabolites in semi solid samples (e.g. intact tissues, tumor biopsies)
Liquid chromatography–tandem mass spectrometry (LC-MS/MS)
LC-MS/MS is a powerful tool for sensitive and specific, quantitative and qualitative analysis of small molecule metabolites. Depending on the type of mass spectrometer, different processes (such as filtration of specific mass-to-charge ratios, collision-induced dissociation or fragmentation, full mass range scans) are available, allowing for accurate quantification in targeted approaches or screening and identification of unknown substances in untargeted setups. Within the Metabolomics Platform, LC-MS/MS-based targeted approaches include analysis of e.g. tricarboxylic acid (TCA) cycle metabolites, biogenic (poly)amines, steroids, catecholamine metabolites and bile acids, as well as untargeted approaches for metabolomic screenings.
Metabolic flux analysis
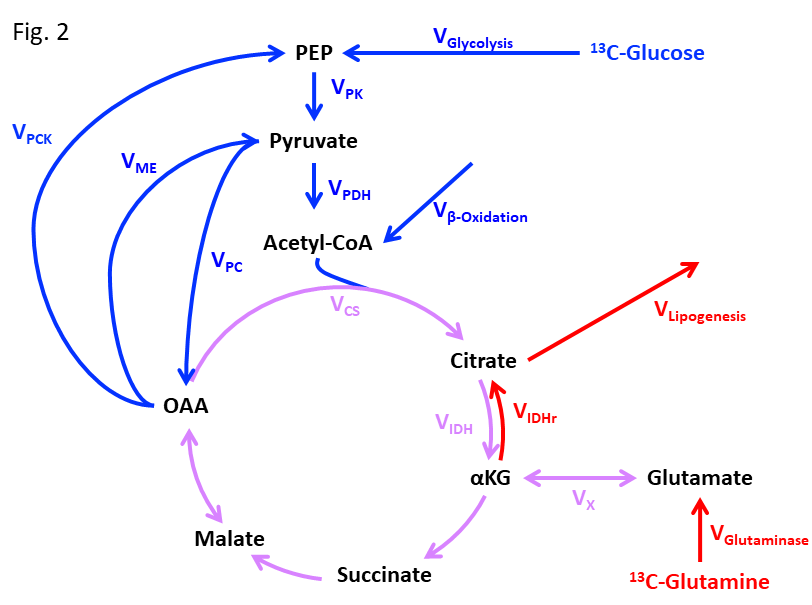
LC-MS/MS methodology is complemented by stable isotope metabolic flux analysis for a more dynamic understanding of the metabolic pathophysiology. Stable isotope metabolic flux analysis can trace the source of oncometabolites, a measure the rate at which they are produced as well as the regulation of such reactions in cell culture models. Depending on the initial tracer used, we can study glycolysis, the TCA cycle (forward and reverse reactions), glucose and fatty acid oxidation and glutamine metabolism – all pathways implicated in cancer pathophysiology (Figure 2).
Services
- NMR spectroscopy analysis in liquid samples and tissues
- LC-MS/MS: Targeted and untargeted profiling of small molecule metabolites in different samples
- Consulting service for setup of experimental designs in translational research projects
- Planning, analysis and interpretation of metabolic flux data
- Methodology/assay setup for translational research projects on request
Methods
NMR spectroscopy
- Metabolite measurement in liquid samples
- HR-MAS for measurement of metabolites in semi-solid samples
Targeted LC-MS/MS
- TCA cycle metabolite profiling (cell culture)
- Steroids (plasma, tissue, cell culture)
- Catecholamine metabolites (plasma, urine, tissue, cell culture)
- Biogenic amines within arginine metabolism (cell culture, serum, urine, feces, tissue)
- Bile acids (plasma, tissue)
- Further methods on request
Untargeted LC-MS/MS-based metabolomics (plasma, cell culture, tissue)
13C-based metabolic flux analysis (cell culture)
Prof. Dr. Triantafyllos Chavakis, Institute for Clinical Chemistry and Laboratory Medicine of the University hospital Carl Gustav Carus Dresden, TU Dresden
Prof. Dr. Peter Mirtschink, Institute for Clinical Chemistry and Laboratory Medicine of the University hospital Carl Gustav Carus Dresden, TU Dresden
Dr. Alexander Funk, Institute for Clinical Chemistry and Laboratory Medicine, University Hospital Carl Gustav Carus, TU Dresden
Email: alexander.funk(at)ukdd.de
Phone: +49 (0)351 458 11319
HEAD, EXPERIMENTAL LC-MS/MS FACILITY
Dr. Mirko Peitzsch, Institute for Clinical Chemistry and Laboratory Medicine, University Hospital Carl Gustav Carus, TU Dresden
Email: Mirko.Peitzsch(at)ukdd.de
Phone: +49 (0)351 458 14578
HEAD, METABOLIC FLUX
Dr. Tiago Alves, Institute for Clinical Chemistry and Laboratory Medicine, University Hospital Carl Gustav Carus, TU Dresden
Email: tiago.alves(at)ukdd.de
Phone: +49 (0)351 458 11707